Data Transformation Explanatory data Analysis
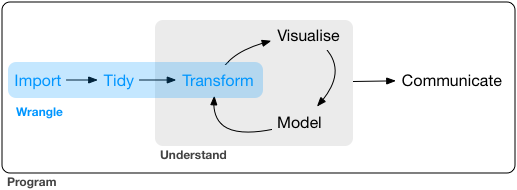
Objectives
Welcome to yet another R blog on data analysis and wranling using R At the end of this tutorial you will:
- understand the role of data wrangling
- understand the basic capabilities of dplyr
- acquire skills to perform common data wrangling using dplyr,stringr,tidyr and forcats packages
What is data wrangling?
Common data wrangling processes
The common data wrangling processes include:
- reducing the size of dataset by selecting certain variables (or columns)
- generating new variable from existing variables
- sorting observation of a variable
- grouping observations based on certain criteria
- reducing variables to groups in order to estimate summary statistic
Use tidyverse functions for achieving this!
already have a blog about these functions
-
dplyr::select()- to select a number of variables from a dataframe -
dplyr::mutate()- to generate a new variable from existing variables -
dplyr::arrange()- to sort observation of a variable -
dplyr::filter()- to group observations that fulfil certain criteria -
dplyr::group_by()anddplyr::summarize()- to reduce variable to groups in order to provide summary statistic
setup
library(tidyverse)
library(lubridate)
new_data <- read_csv("recipe_site_traffic_2212.csv")
dim(new_data)
#> [1] 947 8
names(new_data)
#> [1] "recipe" "calories" "carbohydrate" "sugar" "protein"
#> [6] "category" "servings" "high_traffic"
Take a peek at the recipe site dataset.
The dataset contains:
- 947 observations
- 8 variables
glimpse(new_data)
#> Rows: 947
#> Columns: 8
#> $ recipe <chr> "001", "002", "003", "004", "005", "006", "007", "008", "~
#> $ calories <dbl> NA, 35.48, 914.28, 97.03, 27.05, 691.15, 183.94, 299.14, ~
#> $ carbohydrate <dbl> NA, 38.56, 42.68, 30.56, 1.85, 3.46, 47.95, 3.17, 3.78, 4~
#> $ sugar <dbl> NA, 0.66, 3.09, 38.63, 0.80, 1.65, 9.75, 0.40, 3.37, 3.99~
#> $ protein <dbl> NA, 0.92, 2.88, 0.02, 0.53, 53.93, 46.71, 32.40, 3.79, 11~
#> $ category <chr> "Pork", "Potato", "Breakfast", "Beverages", "Beverages", ~
#> $ servings <chr> "6", "4", "1", "4", "4", "2", "4", "4", "6", "2", "1", "6~
#> $ high_traffic <chr> "High", "High", NA, "High", NA, "High", NA, NA, "High", N~
Next, we examine the first five observations of the data. The rest of the observations are not shown. You can also see the types of variables:
chr(character),int(integer),dbl(double)
new_data|> head(n = 5)
#> # A tibble: 5 x 8
#> recipe calories carbohydrate sugar protein category servings high_traffic
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
#> 1 001 NA NA NA NA Pork 6 High
#> 2 002 35.5 38.6 0.66 0.92 Potato 4 High
#> 3 003 914. 42.7 3.09 2.88 Breakfast 1 <NA>
#> 4 004 97.0 30.6 38.6 0.02 Beverages 4 High
#> 5 005 27.0 1.85 0.8 0.53 Beverages 4 <NA>
Select variables, generate new variable and rename variable
We will work with these functions.
dplyr::select()dplyr::mutate()anddplyr::rename()
Select variables using dplyr::select()
When you work with large datasets with many columns, it is sometimes easier to select only the necessary columns to reduce the dataset size. This is possible by creating a smaller dataset (fewer variables). Then you can work on the initial part of data analysis with this smaller dataset. This will greatly help data exploration.
(new_data <- new_data|>
dplyr::select(everything()))
#> # A tibble: 947 x 8
#> recipe calories carbohydrate sugar protein category servings high_traffic
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
#> 1 001 NA NA NA NA Pork 6 High
#> 2 002 35.5 38.6 0.66 0.92 Potato 4 High
#> 3 003 914. 42.7 3.09 2.88 Breakfast 1 <NA>
#> 4 004 97.0 30.6 38.6 0.02 Beverages 4 High
#> 5 005 27.0 1.85 0.8 0.53 Beverages 4 <NA>
#> 6 006 691. 3.46 1.65 53.9 One Dish Me~ 2 High
#> 7 007 184. 48.0 9.75 46.7 Chicken Bre~ 4 <NA>
#> 8 008 299. 3.17 0.4 32.4 Lunch/Snacks 4 <NA>
#> 9 009 539. 3.78 3.37 3.79 Pork 6 High
#> 10 010 248. 48.5 3.99 114. Chicken 2 <NA>
#> # i 937 more rows
extending select verb
sometimes it is necessary to perform conditional selection on variables because
- at times you need only numerical variables for correlations
- you may only need categorical variables for testing independence
for such a case we can use functions such as
select_if
new_data |> select_if(is.numeric)
#> # A tibble: 947 x 4
#> calories carbohydrate sugar protein
#> <dbl> <dbl> <dbl> <dbl>
#> 1 NA NA NA NA
#> 2 35.5 38.6 0.66 0.92
#> 3 914. 42.7 3.09 2.88
#> 4 97.0 30.6 38.6 0.02
#> 5 27.0 1.85 0.8 0.53
#> 6 691. 3.46 1.65 53.9
#> 7 184. 48.0 9.75 46.7
#> 8 299. 3.17 0.4 32.4
#> 9 539. 3.78 3.37 3.79
#> 10 248. 48.5 3.99 114.
#> # i 937 more rows
the code above will only select variables of class
numeric
Generate new variable using mutate()
With mutate(), you can generate a new variable. For example, in the dataset new_data, we want to create a new variable named log_calories which is a log transformation of calories .
$$log\_calories=\log(calories)$$
And let’s observe the first five observations:
new_data <- new_data|>
dplyr::mutate(log_calories = log(calories))
new_data |>
dplyr::select(log_calories,calories,sugar,category)|>
slice_head(n = 5)
#> # A tibble: 5 x 4
#> log_calories calories sugar category
#> <dbl> <dbl> <dbl> <chr>
#> 1 NA NA NA Pork
#> 2 3.57 35.5 0.66 Potato
#> 3 6.82 914. 3.09 Breakfast
#> 4 4.58 97.0 38.6 Beverages
#> 5 3.30 27.0 0.8 Beverages
extending mutate function
it is often wise to perform conditional mutations on data
sometimes it is necessary to perform conditional mutation on variables such that
- you only mutate is a certain condition is met
we often use
mutate_if(),mutate_atandmutate_allto achieve this
- check data types before the coming operation
map_chr(new_data,class)
#> recipe calories carbohydrate sugar protein category
#> "character" "numeric" "numeric" "numeric" "numeric" "character"
#> servings high_traffic log_calories
#> "character" "character" "numeric"
- we note that category ,servings and high_traffic are characters when in actual fact they should be factors
lets change that
## change characters to factors
new_data <- new_data |>
mutate_if(is.character,as.factor)
## now check the data types
map_chr(new_data,class)
#> recipe calories carbohydrate sugar protein category
#> "factor" "numeric" "numeric" "numeric" "numeric" "factor"
#> servings high_traffic log_calories
#> "factor" "factor" "numeric"
nice!! we have turned every
characterto afactor
Rename variable using rename()
Now, we want to rename
- variable category to
meal_category - variable log_calories to
log_of_calories
(new_data <- new_data |>
rename(meal_category = category,
log_of_calories = log_calories))
#> # A tibble: 947 x 9
#> recipe calories carbohydrate sugar protein meal_category servings
#> <fct> <dbl> <dbl> <dbl> <dbl> <fct> <fct>
#> 1 001 NA NA NA NA Pork 6
#> 2 002 35.5 38.6 0.66 0.92 Potato 4
#> 3 003 914. 42.7 3.09 2.88 Breakfast 1
#> 4 004 97.0 30.6 38.6 0.02 Beverages 4
#> 5 005 27.0 1.85 0.8 0.53 Beverages 4
#> 6 006 691. 3.46 1.65 53.9 One Dish Meal 2
#> 7 007 184. 48.0 9.75 46.7 Chicken Breast 4
#> 8 008 299. 3.17 0.4 32.4 Lunch/Snacks 4
#> 9 009 539. 3.78 3.37 3.79 Pork 6
#> 10 010 248. 48.5 3.99 114. Chicken 2
#> # i 937 more rows
#> # i 2 more variables: high_traffic <fct>, log_of_calories <dbl>
Sorting data and selecting observation
The function arrange() can sort the data. And the function filter()allows you to select observations based on your criteria.
Sorting data using arrange()
We can sort data in ascending or descending order using the arrange() function.
new_data|>
arrange(log_of_calories) |>
relocate(log_of_calories)
#> # A tibble: 947 x 9
#> log_of_calories recipe calories carbohydrate sugar protein meal_category
#> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 -1.97 229 0.14 18.1 11.2 87.3 Lunch/Snacks
#> 2 -1.20 501 0.3 5.19 0.96 1.51 Beverages
#> 3 -0.616 653 0.54 30.6 10.4 0.36 Beverages
#> 4 -0.528 655 0.59 18.8 58 16.4 Dessert
#> 5 -0.446 850 0.64 75.6 3.26 6.95 Breakfast
#> 6 -0.274 277 0.76 1.9 3.76 0.05 Vegetable
#> 7 -0.223 670 0.8 12.3 1.33 6.21 Chicken Breast
#> 8 0.445 588 1.56 59.9 6.57 35 Pork
#> 9 0.761 388 2.14 20.1 0.16 1.42 Beverages
#> 10 1.09 514 2.98 9.81 28.6 19.1 Chicken Breast
#> # i 937 more rows
#> # i 2 more variables: servings <fct>, high_traffic <fct>
- this will like arrange the data based on log_calories from smallest to biggest
- we can arrange from biggest to smallest as well using
desc()
new_data|>
arrange(desc(log_of_calories)) |>
relocate(log_of_calories)
#> # A tibble: 947 x 9
#> log_of_calories recipe calories carbohydrate sugar protein meal_category
#> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 8.20 926 3633. 29.1 0.35 2.32 Chicken
#> 2 7.97 125 2906. 3.52 1.89 179. Pork
#> 3 7.90 227 2703. 6.4 2.17 28.2 Pork
#> 4 7.83 072 2508. 18.1 10.6 84.2 Chicken
#> 5 7.75 908 2332. 7.47 3.62 34.3 One Dish Meal
#> 6 7.73 940 2283. 34.3 5.12 17.6 Chicken Breast
#> 7 7.73 357 2283. 4.5 4.16 31.2 One Dish Meal
#> 8 7.66 056 2122. 26.0 0.52 81.4 Pork
#> 9 7.64 098 2082. 8.09 4.78 28.5 One Dish Meal
#> 10 7.63 782 2068. 34.2 1.46 10.0 Potato
#> # i 937 more rows
#> # i 2 more variables: servings <fct>, high_traffic <fct>
Select observation using filter()
filter() function to select observations based on certain criteria. Here, in this example, we will create a new dataset (which we will name as new_data_m) that contains observations where high_traffic is NA ,in this case NA implies low
new_data_m <- new_data|>
filter(is.na(high_traffic)) |>
relocate(high_traffic)
new_data_m
#> # A tibble: 373 x 9
#> high_traffic recipe calories carbohydrate sugar protein meal_category
#> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 <NA> 003 914. 42.7 3.09 2.88 Breakfast
#> 2 <NA> 005 27.0 1.85 0.8 0.53 Beverages
#> 3 <NA> 007 184. 48.0 9.75 46.7 Chicken Breast
#> 4 <NA> 008 299. 3.17 0.4 32.4 Lunch/Snacks
#> 5 <NA> 010 248. 48.5 3.99 114. Chicken
#> 6 <NA> 011 170. 17.6 4.1 0.91 Beverages
#> 7 <NA> 012 156. 8.27 9.78 11.6 Breakfast
#> 8 <NA> 020 128. 27.6 1.51 8.91 Chicken
#> 9 <NA> 022 40.5 87.9 105. 11.9 Dessert
#> 10 <NA> 023 82.7 3.17 7.95 26.0 Breakfast
#> # i 363 more rows
#> # i 2 more variables: servings <fct>, log_of_calories <dbl>
- we can see that we have a smaller dataset in which all high_traffic observations are
NAValues
Next, we will create a new dataset (named new_data_high_logless0) that contain
high_traffic=='high'andlog_of_calories<0
new_data_high_logless0 <- new_data|>
filter(high_traffic=='High'& log_of_calories<0) |>
relocate(high_traffic,log_of_calories)
new_data_high_logless0
#> # A tibble: 4 x 9
#> high_traffic log_of_calories recipe calories carbohydrate sugar protein
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>
#> 1 High -1.97 229 0.14 18.1 11.2 87.3
#> 2 High -0.274 277 0.76 1.9 3.76 0.05
#> 3 High -0.616 653 0.54 30.6 10.4 0.36
#> 4 High -0.528 655 0.59 18.8 58 16.4
#> # i 2 more variables: meal_category <fct>, servings <fct>
Group data and get summary statistics
Thegroup_by() function allows us to group data based on categorical variable. Using the summarize we do summary statistics for the overall data or for groups created using group_by() function.
Group data using group_by()
The group_by function will prepare the data for group analysis. For example,
- to get summary values for mean
calories, meansugarand meancarbohydrate - for category
new_data_category <- new_data|>
group_by(meal_category)
Summary statistic using summarize()
Now that we have a group data named new_data_category, now, we would summarize our data using the mean and standard deviation (SD) for the groups specified above.
new_data_category|>
summarise(mean_calories = mean(calories, na.rm = TRUE),
mean_sugars = mean(sugar, na.rm = TRUE),
mean_carbohydrate = mean(carbohydrate, na.rm = TRUE))
#> # A tibble: 11 x 4
#> meal_category mean_calories mean_sugars mean_carbohydrate
#> <fct> <dbl> <dbl> <dbl>
#> 1 Beverages 178. 12.5 16.0
#> 2 Breakfast 332. 7.55 39.7
#> 3 Chicken 567. 5.68 30.8
#> 4 Chicken Breast 540. 5.10 21.8
#> 5 Dessert 351. 35.2 55.7
#> 6 Lunch/Snacks 479. 5.31 42.8
#> 7 Meat 585. 5.81 22.2
#> 8 One Dish Meal 579. 6.01 50.4
#> 9 Pork 630. 8.04 28.1
#> 10 Potato 425. 3.72 58.2
#> 11 Vegetable 245. 5.07 23.7
To calculate the frequencies for two variables for the recipe dataset
- category
- high_traffic
new_data|>
group_by(meal_category)|>
count(high_traffic, sort = TRUE)
#> # A tibble: 22 x 3
#> # Groups: meal_category [11]
#> meal_category high_traffic n
#> <fct> <fct> <int>
#> 1 Beverages <NA> 87
#> 2 Potato High 83
#> 3 Vegetable High 82
#> 4 Pork High 77
#> 5 Breakfast <NA> 73
#> 6 Meat High 59
#> 7 Lunch/Snacks High 57
#> 8 Dessert High 53
#> 9 Chicken Breast <NA> 52
#> 10 One Dish Meal High 52
#> # i 12 more rows
or
new_data |>
count(meal_category, high_traffic, sort = TRUE)
#> # A tibble: 22 x 3
#> meal_category high_traffic n
#> <fct> <fct> <int>
#> 1 Beverages <NA> 87
#> 2 Potato High 83
#> 3 Vegetable High 82
#> 4 Pork High 77
#> 5 Breakfast <NA> 73
#> 6 Meat High 59
#> 7 Lunch/Snacks High 57
#> 8 Dessert High 53
#> 9 Chicken Breast <NA> 52
#> 10 One Dish Meal High 52
#> # i 12 more rows
More complicated dplyr verbs
To be more efficient, use multiple dplyr functions in one line of R code. For example,
new_data |>
filter(meal_category != "Pork", calories>10, !is.na(protein))|>
dplyr::select(recipe,meal_category,calories, sugar,carbohydrate,protein, high_traffic)|>
mutate(meal_category=if_else(is.na(meal_category),"low",meal_category)) |>
group_by(meal_category)|>
summarize(mean_calories = mean(calories, na.rm = TRUE),
mean_sugars = mean(sugar, na.rm = TRUE),
mean_carbohydrate = mean(carbohydrate, na.rm = TRUE),
freq = n())
#> # A tibble: 10 x 5
#> meal_category mean_calories mean_sugars mean_carbohydrate freq
#> <chr> <dbl> <dbl> <dbl> <int>
#> 1 Beverages 186. 12.9 16.0 88
#> 2 Breakfast 338. 7.50 39.3 104
#> 3 Chicken 567. 5.68 30.8 69
#> 4 Chicken Breast 552. 4.88 22.1 92
#> 5 Dessert 361. 35.1 53.7 75
#> 6 Lunch/Snacks 497. 5.29 42.8 79
#> 7 Meat 601. 5.44 22.0 72
#> 8 One Dish Meal 579. 6.01 50.4 67
#> 9 Potato 430. 3.66 58.6 82
#> 10 Vegetable 255. 4.99 24.0 75
Data transformation for categorical variables
forcats package
Data transformation for categorical variables (factor variables) can be facilitated using the forcats package.
Conversion from numeric to factor variables
Now, we will convert the integer (numerical) variable to a factor (categorical) variable. For example, we will generate a new factor (categorical) variable named high_sugars from variables sugars (both double variables). We will label high_bpas High or Not High.
The criteria:
- if sugar
\(sugar \geq 20 or is.na\)then labelled as High, else is Not High
new_data <- new_data|>
mutate(high_sugar = if_else(sugar >= 20|is.na(sugar) ,
"High", "Not High"))
new_data|> count(high_sugar)
#> # A tibble: 2 x 2
#> high_sugar n
#> <chr> <int>
#> 1 High 134
#> 2 Not High 813
of by using cut()
new_data <- new_data|>
filter(!is.na(carbohydrate)) |>
mutate(cat_carboydrates = cut(carbohydrate, breaks = c(-Inf, 120, 130, Inf),
labels = c('<120', '121-130', '>130')))
new_data|> count(cat_carboydrates)
#> # A tibble: 3 x 2
#> cat_carboydrates n
#> <fct> <int>
#> 1 <120 852
#> 2 121-130 6
#> 3 >130 37
new_data|>
group_by(cat_carboydrates)|>
summarize(min_carbohydrates = min(carbohydrate),
max_carbohydrates = max(carbohydrate))
#> # A tibble: 3 x 3
#> cat_carboydrates min_carbohydrates max_carbohydrates
#> <fct> <dbl> <dbl>
#> 1 <120 0.03 120.
#> 2 121-130 124. 128.
#> 3 >130 132. 530.
Recoding variables
We use this function to recode variables from old to new levels. For example:
new_data <- new_data|>
mutate(cat_carboydrates_new = recode(cat_carboydrates, "<120" = "120 or less",
"121-130" = "121 to 130",
">130" = "131 or higher"))
new_data|> count(cat_carboydrates_new)
#> # A tibble: 3 x 2
#> cat_carboydrates_new n
#> <fct> <int>
#> 1 120 or less 852
#> 2 121 to 130 6
#> 3 131 or higher 37
Changing the level of categorical variable
Variable cat_carboydrates_new will be ordered as
- less or 120, then
- 121 - 130, then
- 131 or higher
levels(new_data$cat_carboydrates_new)
#> [1] "120 or less" "121 to 130" "131 or higher"
new_data|> count(cat_carboydrates_new)
#> # A tibble: 3 x 2
#> cat_carboydrates_new n
#> <fct> <int>
#> 1 120 or less 852
#> 2 121 to 130 6
#> 3 131 or higher 37
To change the order (in reverse for example), we can use fct_relevel. Below the first level group is sbp above 130, followed by 121 to 130 and the highest group is less than 120.
new_data <- new_data|>
mutate(relevel_cat_carboydrates = fct_relevel(cat_carboydrates, ">130", "121-130", "<120"))
levels(new_data$relevel_cat_carboydrates)
#> [1] ">130" "121-130" "<120"
new_data|> count(relevel_cat_carboydrates)
#> # A tibble: 3 x 2
#> relevel_cat_carboydrates n
#> <fct> <int>
#> 1 >130 37
#> 2 121-130 6
#> 3 <120 852